Accelerating Computational Chemistry Research with the Cloud
Project phases
- Phase 1 - Accelerating Computational Chemistry Research with the Cloud (current page)
- Phase 2 - Accelerating Computational Chemistry Research with the Cloud Phase II
To support and enable complex research computations, faculty members who conduct research in Computational Chemistry at the University of British Columbia (UBC), in collaboration with the UBC Cloud Innovation Centre (UBC CIC), developed a solution that moves the existing program into the Amazon Web Services (AWS) cloud for greater control, flexibility, and efficiency.
Approach
To understand and predict molecular behaviours, Computational Chemistry researchers have to run complex calculations and computer simulations. The current process is complicated, time-consuming and requires manual operation and maintenance.
The solution aims to give Computational Chemistry researchers the ability to control and optimize computing resources when running their calculations in the AWS cloud so that the research process is expedited. The existing program used by the researchers requires long periods of time, sometimes days, for some calculations to run and is manually maintained by one person, so the time to execute calculations or fix issues that arise is prolonged. By integrating the existing program with AWS services, program calculations can be divided to run at the same time and users can see the status of their calculations to determine if it will run, fail, or if they would like to abort. This gives researchers more flexibility and management over how and when calculations are run.
Supporting Artifacts
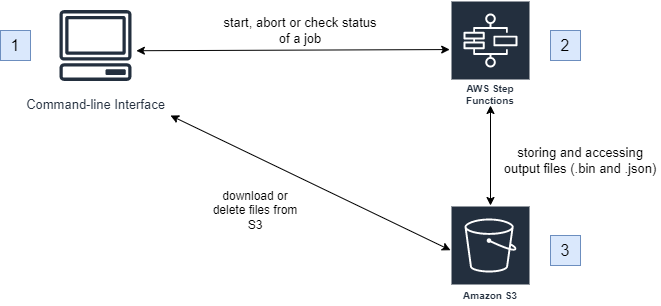
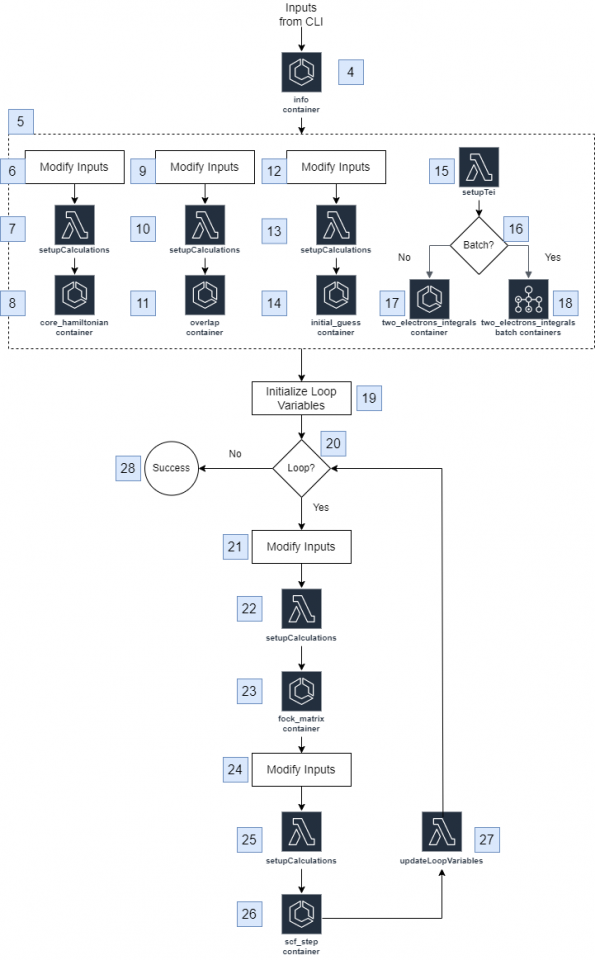
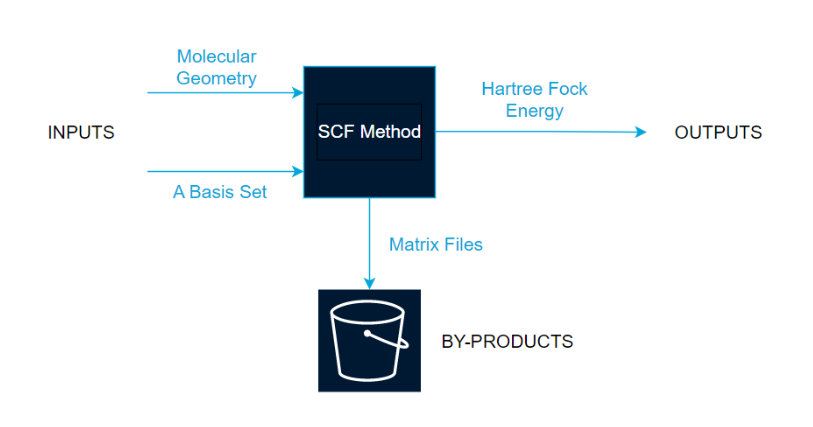
Architecture Diagram
Technical Details
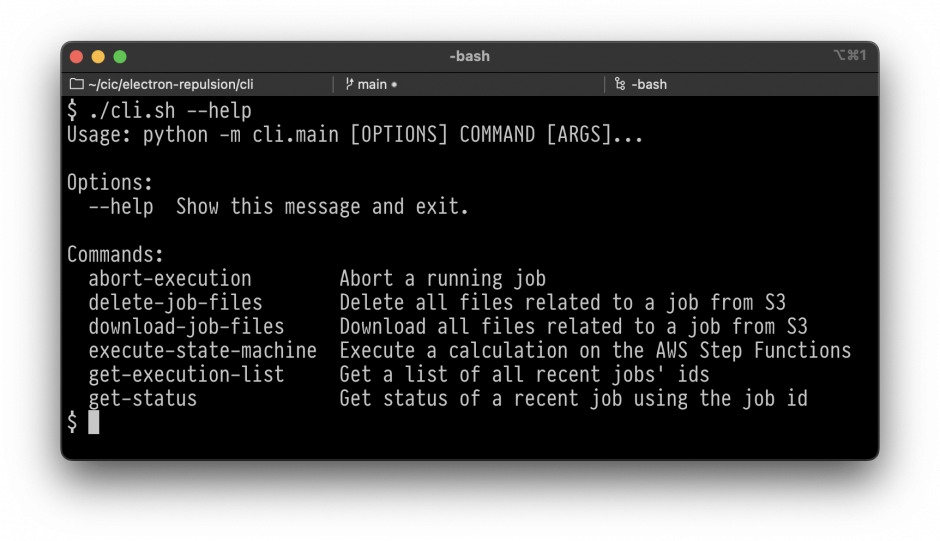
Users interact with the solution using a command-line interface (CLI) that supports specific inputs related to calculation jobs. These inputs allow users to do the following:
- execute a job,
- check the status of a job,
- list all jobs and their statuses,
- abort a job that is running,
- delete all the files related to a job ID from the S3 bucket, and
- download all the files related to a job ID to the local computer running the CLI.
These inputs trigger the step functions that run the calculations, which in turn access an Amazon S3 Bucket to store and pull .bin and .json files for the calculations.
The user will enter an input in the CLI and the solution first executes an AWS Elastic Container Service (ECS) task to run an info step so that the result can be stored in the S3 bucket as a .json file. From there, four calculations are run in parallel: the core_hamiltonian step, the overlap step, the initial_guess step, and the two_electrons_integrals step. The core_hamiltonian step, the overlap step, and the initial_guess step follow similar processes where a name is added to the input, a setupCalculations Lambda function sets up the parameters to run the step, and an ECS task is run to execute the step, which generates a standard output and a calculation result to store as a .json file and a .bin file respectively in the S3 bucket.
The two_electron_integrals step follows a different process which uses the info step result to get the calculation’s basis_set_instance_size value and prepares to split the calculation into parts using the numSlices value, which can be specified by the user in the CLI or automatically. The program determines if AWS Batch will be used to complete the calculation. If it is not used, the step runs as an ECS task to calculate integrals within a range using the basis_set_instance_size value; if it is used, a .txt file generated during the setup distributes tasks to the batch jobs and each job outputs a .bin file and a .json file to the S3 bucket.
The inputs have a loopData dictionary added to track the number of iterations calculated. The loop will terminate once a number of iterations has reached a limit or the value of the hartree_fock_energy falls below a threshold value, both of which can be specified in the CLI or use a default value. The fock_matrix step and the scf_step are run using the same process as the core_hamiltonian step, the overlap step, and the initial_guess step, with standard outputs stored as .json files and calculation results stored as .bin files in the S3 bucket. The loopData dictionary updates, and once the maximum number of interactions or the minimum value of hartree_diff is reached, the loop terminates and the execution is marked as successful.
Link to solution on GitHub: https://github.com/UBC-CIC/electron-repulsion/tree/phase1
UI SCREENSHOT
Acknowledgements
Chen Greif
Ury Segal
Mark Thachuk
Alexandra Bunger
Photo by: Yuri Shkoda on pexels.com
About the University of British Columbia Cloud Innovation Centre (UBC CIC)
UBC’s CIC is a public-private collaboration between UBC and Amazon. A CIC identifies digital transformation challenges, the problems or opportunities that matter to the community, and provides subject matter expertise and CIC leadership.
Using Amazon’s innovation methodology, dedicated UBC and Amazon CIC staff will work with students, staff and faculty, as well as community, government or not-for-profit organizations to define challenges, to engage with subject matter experts, to identify a solution, and to build a Proof of Concept (PoC). Through co-op and work-integrated learning, students also have an opportunity to learn new skills which they will later be able to apply in the workforce.
UBC’s CIC focuses on Community Health and Wellbeing.

