Serratus: Ultra-high throughput discovery of new coronaviruses
Project phases
- Phase 1 - Serratus: Ultra-high throughput discovery of new coronaviruses (current page)
- Phase 2 - Serratus: Ultra-high throughput discovery of new coronaviruses – Phase 2
An Open Science project for ultra-rapid discovery of known and unknown coronaviruses in response to the COVID-19 pandemic by screening all public RNA-sequencing data.
SARS-COV-2 and the ongoing COVID-19 pandemic
The COVID-19 pandemic is caused by the SARS-CoV-2 coronavirus which in late 2019 jumped species likely from bats and into humans. SARS-CoV-2 is one coronavirus, and there are dozens of species that infect diverse animals across the world.
It is critically important to catalogue all coronaviruses and their animal reservoirs, since coronaviruses can mix RNA (recombine) resulting in new viral strains, and potentially new outbreaks. Since SARS-CoV-2 is a novel virus, it is of paramount importance to identify related viruses as they are potential sources for recombination.
The NCBI Short Read Archive (SRA) contains sequencing data from 3+ million biological samples, or over 4,000,000 gigabytes (4PB) of raw DNA/RNA sequence data. This covers thousands of animal species across all geographic regions.
Serratus is an ultra-rapid discovery project to identify new species of coronavirus (CoV) by analyzing all public sequencing data, including all of the SRA. Leveraging the massive HPC capabilities on AWS cloud we are performing an ultra-deep search for novel viruses to help fight the pandemic.
OPEN SCIENCE AND OPEN DATA
Our primary goal is to generate the coronavirus data to accelerate the global research efforts in fighting SARS-CoV-2. This means sharing all data and tools immediately.
We adhere to the Bermuda Principles set out originally by the Human Genome Project, all data is freely and publicly available within 24 hours of generation. If there is a way CoV sequence data can assist your research, please reach out and we can work towards advancing COVID-19 related applications.
JOIN THE SERRATUS COLLABORATION!
We are actively looking for collaborators to join Serratus and help transform this data into meaningful and actionable information in the fight against COVID-19.
We believe that it is through open-collaboration and data-sharing that coronavirus research will progress most rapidly to help our communities.
- Computational Virology (discovery, annotation, …)
- Coronavirus Phylogenetics
- Virus curation and taxonomy
- Viral ecology and population modelling
- Bioinformatic algorithm development (alignment, homology search, …)
- High-dimensional data exploration and machine learning
- R package development
- Web-data interfacing
- SQL Database Admins
REQUEST A FREE VIRAL REPORT WITH YOUR SEQUENCING DATA. TOMORROW.
Serratus is hyper-optimized to rapidly analyze huge sequencing datasets with a throughput of over 1 Million sequencing libraries analyzed per day.
If you have access to a large sets of RNA-seq, meta-genomic or meta-transcriptomic sequencing data, we are offering to process this data at no cost within 24 hours of upload. You will receive a detailed report on the presence of vertebrate viruses in your samples and the associated alignment files.
We only ask that adequate meta-data is provided and the reports and viral assemblies from CoV+ samples be shared publicly.
High priority samples:
- Any sequencing data from bats or bat environments
- Sequencing data from mammals with known or unknown respiratory disease
- Samples taken from immuno-compromised hosts
- Samples taken from hosts with suspected viral infection
IMPACT OF SERRATUS
Expanding the known repertoire of coronaviruses will not only help determine the origins of this pandemic, it can help prevent another one. Serratus can directly help fight the COVID-19 pandemic by:
- Identification of the natural animal reservoirs of different coronaviruses and high-risk sources of viral recombination
- Improving the specificity of SARS-CoV-2 diagnostic tests through the identification of potential false-positive or cross-reactive viruses
- Improvement of SARS-CoV-2 vaccine development by providing rich evolutionary data to understand how viral surface proteins change over time
Architecture Diagram
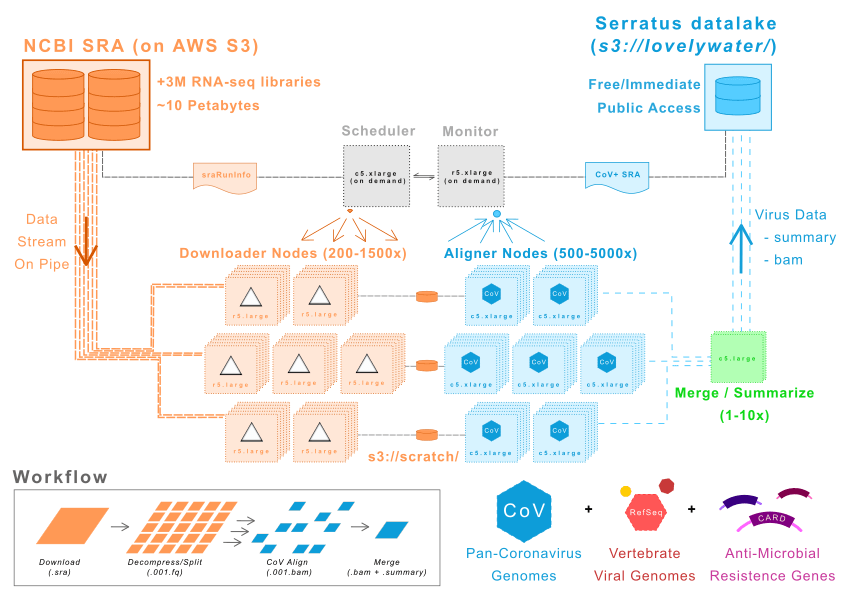
UNDER THE HOOD
In February 2020, AWS mirrored the NCBI Short Read Archive (SRA) onto their S3 servers as an Open Data-set which allows for an unprecedented rate of access to the petabytes of raw data.
To perform the ultra-high throughput CoV search, AWS cloud computing was employed to create a 22,500 vCPU HPC cluster (1460 x R5.xlarge, 4120 x C5.xlarge, and 90 x C5.large EC2 instances). Using this hyper-parallelized architecture we could bypass conventional networking and disk IO limitations that would limit a conventional cluster and get our scientific results faster.
The Serratus architecture is cost-optimized for big data analysis, achieving a maximum alignment throughput of over 1 Million sequencing libraries per day, at a cost of only $0.01 per library.
EARLY RESULTS THAT WE CAN SHARE
Serratus is an ongoing research project and we will post updates on the site as they become available. If you would like more information or would like to get involved with this project please contact us github.
Serratus: The ultra-deep search to discover novel coronaviruses.
Acknowledgements
SERRATUS ALIGNMENT TEAM
- Artem Babaian – University of British Columbia
- Robert C. Edgar – unaffiliated
- Jeff Taylor – unaffiliated
- Kyl Wellman – unaffiliated
SERRATUS ASSEMBLY TEAM
- Rayan Chikhi – Institut Pasteur & CNRS
- Anton Korobeynikov – Center for Algorithmic Biotechnologies, Saint Petersburg State University
- Ryan J. McLaughlin – University of British Columbia
- Dmitry Meleshko – Center for Algorithmic Biotechnologies, Saint Petersburg State University
SERRATUS ANNOTATION TEAM
- Tomer Altman – Altman Analytics LLC and University of British Columbia (BCB2)
- Steven J. Hallam – University of British Columbia
SERRATUS WEB/WIKI TEAM
- Victor Lin – unaffiliated
- Dan Lohr – unaffiliated
TANTALUS TEAM
- Gherman Novakovsky – University of British Columbia
About the University of British Columbia Cloud Innovation Centre (UBC CIC)
The UBC CIC is a public-private collaboration between UBC and Amazon Web Services (AWS). A CIC identifies digital transformation challenges, the problems or opportunities that matter to the community, and provides subject matter expertise and CIC leadership.
Using Amazon’s innovation methodology, dedicated UBC and AWS CIC staff work with students, staff and faculty, as well as community, government or not-for-profit organizations to define challenges, to engage with subject matter experts, to identify a solution, and to build a Proof of Concept (PoC). Through co-op and work-integrated learning, students also have an opportunity to learn new skills which they will later be able to apply in the workforce.