Serratus: Ultra-high throughput discovery of new coronaviruses – Phase 2
Project phases
- Phase 1 - Serratus: Ultra-high throughput discovery of new coronaviruses
- Phase 2 - Serratus: Ultra-high throughput discovery of new coronaviruses – Phase 2 (current page)
Serratus is an Open Science project for ultra-rapid discovery of known and unknown coronavirus in response to the COVID-19 pandemic.
For the past decade scientists across the world have shared DNA- and RNA-sequencing data in public repositories. Today, this amounts to an exponentially growing 50,000,000+ gigabytes of raw data. These collections contain vast biological diversity, from deep ice-core samples in the Antarctic, to tropical birds in Madagascar, to experimental cells in a lab in Vancouver. Viruses are also present in these samples, but unless specifically searched for, they can go undiscovered.
To help fight the COVID-19 pandemic caused by the SARS-CoV-2 coronavirus, an international team of scientists led by researchers at the University of British Columbia re-analyzed public sequences databases of 3.84 million samples, or 5.6 quadrillion (5,620,086,903,602,832) nucleotides to catalogue every known, and unknown coronaviruses. This not only can help explain the origins of the SARS-CoV-2 virus, it can help scientists and policy makers decide where to focus future research and monitoring to prevent future pandemics.
With standard high-performance computing clusters this analysis would normally take over a year to compute. To uncover the global biodiversity of coronaviruses today, when the results can mitigate the impact of this pandemic, we created Serratus.
Serratus is an ultra-high throughput sequence alignment platform, based off of Amazon Web Services. Serratus can analyze 1,000,000+ sequencing samples per day for under 1 cent per sample with a dynamic-scaling EC2 cluster reaching 22,250 simultaneous vCPU. Leveraging AWS systems, we could perform 713 years of computing in the span of 7 days. Ultra-high throughput alignment at this efficiency will radically change computational biology in the years to come, starting with virology.
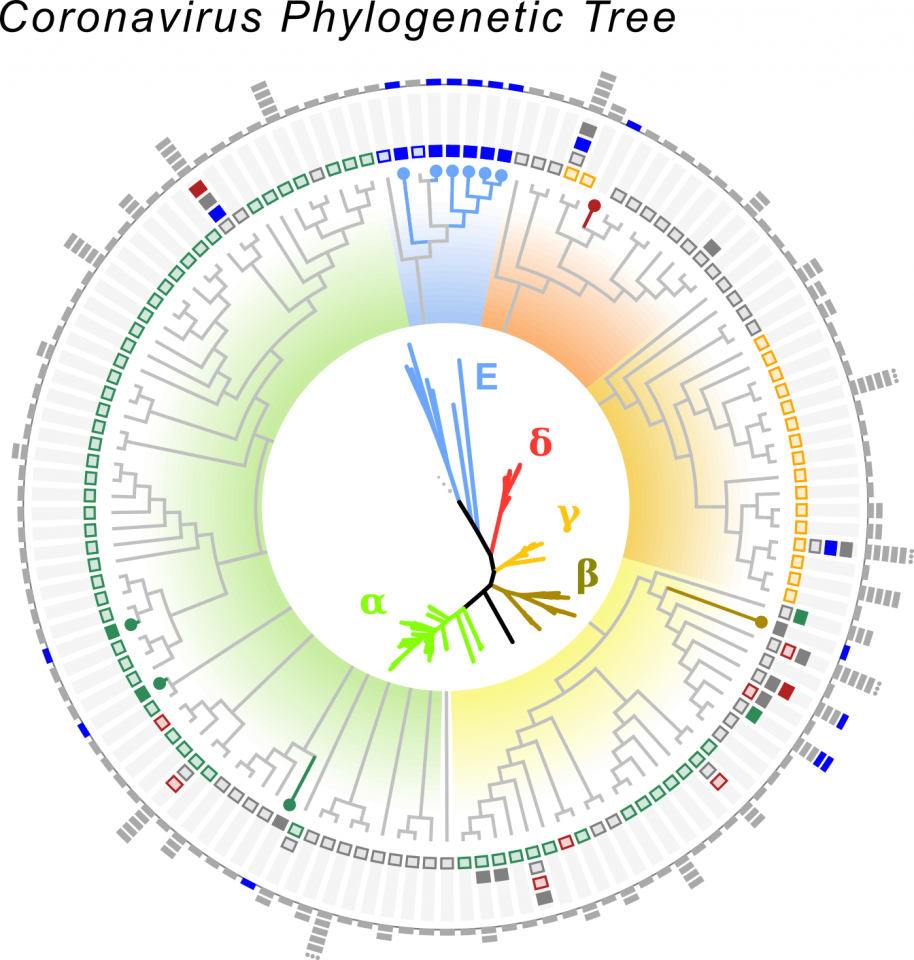
CORONA- AND CORONA-LIKE VIRUS PHYLOGENETIC TREE.
We identified >50,000 samples containing Coronaviruses, from which we recovered >11,000 intact viral sequences. There are four major groupings of Coronaviruses called alpha, beta, gamma and delta. With Serratus we identified a new group of Corona-like viruses found in aquatic species like seahorse, axolotl and fugu fish. These viruses challenge the textbook definition of what a Coronavirus can be like and give us insights into the evolutionary origins of these viruses, to learn more, read the Serratus pre-print.
IMPACT OF SERRATUS
Expanding the known repertoire of coronaviruses will not only help determine the origins of this pandemic, it can help prevent another one. Serratus can directly help fight the COVID-19 pandemic by:
- Releasing over 11,000 coronavirus complete and partial genomes to expand the known CoV biodiversity
- Improving SARS-CoV-2 vaccine development by providing rich evolutionary data to understand how viral surface proteins change over time
- Discovering a new group of corona-like viral species which gives us the deepest insight into the evolutionary origins of coronaviruses
To better prepare us for the next pandemics we have created the world’s most comprehensive viral discovery resource for all viruses that can potentially infect humans
- Viral index of 3.84 million samples to aid the identification of viruses and their animal resources
OPEN SCIENCE AND OPEN DATA
Our goal is to identify every available viral sequence to assist global research efforts in fighting COVID-19 today, and help us prepare and mitigate the next pandemic.
We adhere to the Bermuda Principles set out originally by the Human Genome Project, all code and data are freely and publicly available within 24 hours of generation. If there is a way CoV sequence data can assist your research, please reach out and we can work towards advancing COVID-19 related applications.
– or –
SERRATUS IS AN OPEN COLLABORATION
We are actively looking for collaborators to join Serratus and help transform this data into meaningful and actionable information in the fight against COVID-19, and to fortify our response to future pandemics.
We believe that it is through open-collaboration and data-sharing that coronavirus research will progress most rapidly. Specific expertise sought includes:
- Computational Virology
- Coronavirus Phylogenetics
- Virus curation and taxonomy
- Viral ecology and population modelling
- Bioinformatic algorithm development (alignment, homology search, …)
- High-dimensional data exploration and machine learning
- Web and database interfaces (JavaScript, PostgreSQL, R/Shiny …)
About the University of British Columbia Cloud Innovation Centre (UBC CIC)
The UBC CIC is a public-private collaboration between UBC and Amazon Web Services (AWS). A CIC identifies digital transformation challenges, the problems or opportunities that matter to the community, and provides subject matter expertise and CIC leadership.
Using Amazon’s innovation methodology, dedicated UBC and AWS CIC staff work with students, staff and faculty, as well as community, government or not-for-profit organizations to define challenges, to engage with subject matter experts, to identify a solution, and to build a Proof of Concept (PoC). Through co-op and work-integrated learning, students also have an opportunity to learn new skills which they will later be able to apply in the workforce.

